Selected publications
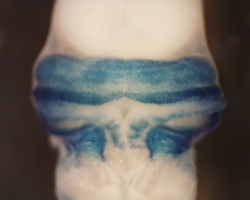
math1 &
development of the cerebellum
development of the cerebellum
Ben-Arie N, McCall A E, Berkman S, Eichele G, Bellen H J, and Zoghbi H Y.
Evolutionary conservation of sequence and expression of the bHLH protein Atonal suggests a conserved role in neurogenesis.
Hum Mol Genet 5: 1207-1216 (1996). http://www.ncbi.nlm.nih.gov/pubmed/8872459
Ben-Arie N, Bellen H J, Armstrong D L, McCall A E, Gordadze P R, Guo Q, Matzuk M M, and Zoghbi H Y.
Math1 is essential for genesis of cerebellar granule neurons.
Nature 390: 169-172 (1997). http://www.ncbi.nlm.nih.gov/pubmed/9367153
Bermingham N A, Hassan B A, Price S D, Vollrath M A, Ben-Arie N, Eatock R A, Bellen H J, Lysakowski A, and Zoghbi H Y.
Math1: an essential gene for the generation of inner ear hair cells.
Science 284: 1837-1841 (1999). http://www.ncbi.nlm.nih.gov/pubmed/10364557
Ben-Arie N, Hassan B A, Bermingham N A, Malicki D M, Armstrong D, Matzuk M, Bellen H J, and Zoghbi H Y.
Functional conservation of atonal and Math1 in the CNS and PNS.
Development 127: 1039-1048 (2000). http://www.ncbi.nlm.nih.gov/pubmed/10662643
Helms A W, Abney A L, Ben-Arie N, Zoghbi H Y, and Johnson J E.
Autoregulation and multiple enhancers control Math1 expression in the developing nervous system.
Development 127: 1185-1196 (2000). http://www.ncbi.nlm.nih.gov/pubmed/10683172
Meshorer E, Erb C, Gazit R, Pavlovsky L, Kaufer D, Friedman A, Glick D, Ben-Arie N, and Soreq H.
Alternative splicing and neuritic mRNA translocation under long-term neuronal hypersensitivity.
Science 295: 508-512 (2002). http://www.ncbi.nlm.nih.gov/pubmed/11799248
Canzoniere D, Farioli-Vecchioli S, Conti F, Ciotti M T, Tata A M, Augusti-Tocco G, Mattei E, Lakshmana M K, Krizhanovsky V, Reeves S A, Giovannoni R, Castano F, Servadio A, Ben-Arie N, and Tirone F.
Dual control of neurogenesis by PC3 through cell cycle inhibition and induction of Math1.
J Neurosci 24: 3355-3369 (2004). http://www.ncbi.nlm.nih.gov/pubmed/15056715
Gazit R, Krizhanovsky V, and Ben-Arie N.
Math1 controls cerebellar granule cell differentiation by regulating multiple components of the Notch signaling pathway.
Development 131: 903-913 (2004). http://www.ncbi.nlm.nih.gov/pubmed/14757642
Krizhanovsky V and Ben-Arie N.
A novel role for the choroid plexus in BMP-mediated inhibition of differentiation of cerebellar neural progenitors.
Mech Dev 123: 67-75 (2006). http://www.ncbi.nlm.nih.gov/pubmed/16325379
Krizhanovsky V, Soreq L, Kliminski V, and Ben-Arie N.
Math1 target genes are enriched with evolutionarily conserved clustered E-box binding sites.
J Mol Neurosci 28: 211-229 (2006). http://www.ncbi.nlm.nih.gov/pubmed/16679559
Evolutionary conservation of sequence and expression of the bHLH protein Atonal suggests a conserved role in neurogenesis.
Hum Mol Genet 5: 1207-1216 (1996). http://www.ncbi.nlm.nih.gov/pubmed/8872459
Ben-Arie N, Bellen H J, Armstrong D L, McCall A E, Gordadze P R, Guo Q, Matzuk M M, and Zoghbi H Y.
Math1 is essential for genesis of cerebellar granule neurons.
Nature 390: 169-172 (1997). http://www.ncbi.nlm.nih.gov/pubmed/9367153
Bermingham N A, Hassan B A, Price S D, Vollrath M A, Ben-Arie N, Eatock R A, Bellen H J, Lysakowski A, and Zoghbi H Y.
Math1: an essential gene for the generation of inner ear hair cells.
Science 284: 1837-1841 (1999). http://www.ncbi.nlm.nih.gov/pubmed/10364557
Ben-Arie N, Hassan B A, Bermingham N A, Malicki D M, Armstrong D, Matzuk M, Bellen H J, and Zoghbi H Y.
Functional conservation of atonal and Math1 in the CNS and PNS.
Development 127: 1039-1048 (2000). http://www.ncbi.nlm.nih.gov/pubmed/10662643
Helms A W, Abney A L, Ben-Arie N, Zoghbi H Y, and Johnson J E.
Autoregulation and multiple enhancers control Math1 expression in the developing nervous system.
Development 127: 1185-1196 (2000). http://www.ncbi.nlm.nih.gov/pubmed/10683172
Meshorer E, Erb C, Gazit R, Pavlovsky L, Kaufer D, Friedman A, Glick D, Ben-Arie N, and Soreq H.
Alternative splicing and neuritic mRNA translocation under long-term neuronal hypersensitivity.
Science 295: 508-512 (2002). http://www.ncbi.nlm.nih.gov/pubmed/11799248
Canzoniere D, Farioli-Vecchioli S, Conti F, Ciotti M T, Tata A M, Augusti-Tocco G, Mattei E, Lakshmana M K, Krizhanovsky V, Reeves S A, Giovannoni R, Castano F, Servadio A, Ben-Arie N, and Tirone F.
Dual control of neurogenesis by PC3 through cell cycle inhibition and induction of Math1.
J Neurosci 24: 3355-3369 (2004). http://www.ncbi.nlm.nih.gov/pubmed/15056715
Gazit R, Krizhanovsky V, and Ben-Arie N.
Math1 controls cerebellar granule cell differentiation by regulating multiple components of the Notch signaling pathway.
Development 131: 903-913 (2004). http://www.ncbi.nlm.nih.gov/pubmed/14757642
Krizhanovsky V and Ben-Arie N.
A novel role for the choroid plexus in BMP-mediated inhibition of differentiation of cerebellar neural progenitors.
Mech Dev 123: 67-75 (2006). http://www.ncbi.nlm.nih.gov/pubmed/16325379
Krizhanovsky V, Soreq L, Kliminski V, and Ben-Arie N.
Math1 target genes are enriched with evolutionarily conserved clustered E-box binding sites.
J Mol Neurosci 28: 211-229 (2006). http://www.ncbi.nlm.nih.gov/pubmed/16679559
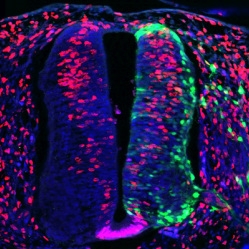
NATO3 &
DEVELOPMENT OF THE SPINAL CORD AND MID-BRAIN
DEVELOPMENT OF THE SPINAL CORD AND MID-BRAIN
Segev E, Halachmi N, Salzberg A, and Ben-Arie N.
Nato3 is an evolutionarily conserved bHLH transcription factor expressed in the CNS of Drosophila and mouse.
Mech Dev 106: 197-202 (2001). http://www.ncbi.nlm.nih.gov/pubmed/11472856
HTML PDF
Mansour A A, Nissim-Eliraz E, Zisman S, Golan-Lev T, Schatz O, Klar A, and Ben-Arie N.
Foxa2 regulates the expression of Nato3 in the floor plate by a novel evolutionarily conserved promoter.
Mol Cell Neurosci 46: 187-199 (2011). http://www.ncbi.nlm.nih.gov/pubmed/20849957
Nissim-Eliraz E, Zisman S, Schatz O, and Ben-Arie N.
Nato3 integrates with the Shh-Foxa2 transcriptional network regulating the differentiation of midbrain dopaminergic neurons.
J Mol Neurosci 51: 13-27 (2013). http://www.ncbi.nlm.nih.gov/pubmed/23254923
Mansour A A, Khazanov-Zisman S, Netser Y, Klar A, and Ben-Arie N.
Nato3 plays an integral role in dorsoventral patterning of the spinal cord by segregating floor plate/p3 fates via Nkx2.2 suppression and Foxa2 maintenance.
Development 141: 574-584 (2014). http://www.ncbi.nlm.nih.gov/pubmed/24401371
HTML PDF
Khazanov S, Paz Y, Hefetz A, Gonzales BJ, Netser Y, Mansour AA, and Ben-Arie N
Uncovering floor plate descendants in the ependyma of adult mouse CNS using mapping of Nato3-expressing cells.
Int. J. Devel. Biol. (doi: 10.1387/ijdb.160232nb). www.ncbi.nlm.nih.gov/pubmed/27528042
HTML PDF
Nato3 is an evolutionarily conserved bHLH transcription factor expressed in the CNS of Drosophila and mouse.
Mech Dev 106: 197-202 (2001). http://www.ncbi.nlm.nih.gov/pubmed/11472856
HTML PDF
Mansour A A, Nissim-Eliraz E, Zisman S, Golan-Lev T, Schatz O, Klar A, and Ben-Arie N.
Foxa2 regulates the expression of Nato3 in the floor plate by a novel evolutionarily conserved promoter.
Mol Cell Neurosci 46: 187-199 (2011). http://www.ncbi.nlm.nih.gov/pubmed/20849957
Nissim-Eliraz E, Zisman S, Schatz O, and Ben-Arie N.
Nato3 integrates with the Shh-Foxa2 transcriptional network regulating the differentiation of midbrain dopaminergic neurons.
J Mol Neurosci 51: 13-27 (2013). http://www.ncbi.nlm.nih.gov/pubmed/23254923
Mansour A A, Khazanov-Zisman S, Netser Y, Klar A, and Ben-Arie N.
Nato3 plays an integral role in dorsoventral patterning of the spinal cord by segregating floor plate/p3 fates via Nkx2.2 suppression and Foxa2 maintenance.
Development 141: 574-584 (2014). http://www.ncbi.nlm.nih.gov/pubmed/24401371
HTML PDF
Khazanov S, Paz Y, Hefetz A, Gonzales BJ, Netser Y, Mansour AA, and Ben-Arie N
Uncovering floor plate descendants in the ependyma of adult mouse CNS using mapping of Nato3-expressing cells.
Int. J. Devel. Biol. (doi: 10.1387/ijdb.160232nb). www.ncbi.nlm.nih.gov/pubmed/27528042
HTML PDF

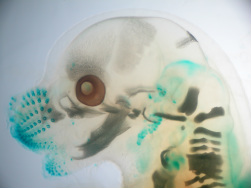
FINGERIN &
DEVELOPMENT OF THE DRG, LIMBS AND FINGERS
DEVELOPMENT OF THE DRG, LIMBS AND FINGERS
Schatz O, Langer E, and Ben-Arie N.
Gene dosage of the transcription factor Fingerin (bHLHA9) affects digit development and links syndactyly to ectrodactyly.
Human Mol. Genetics (2014). doi: 10.1093/hmg/ddu257.
HTML PDF
Bohic M, Marics I, De Pais Paiva Santos C, Ben-Arie N, Malapert P, Salio C, Reynders A, Le Feuvre Y, Saurin AJ, and Moqrich A.
Loss of bhlha9 impairs thermotaxis and formalin-evoked pain in a sexually dimorphic manner.
Cell Reports 30 (3): 602-610.e6 (2020).
HTML PDF
Gene dosage of the transcription factor Fingerin (bHLHA9) affects digit development and links syndactyly to ectrodactyly.
Human Mol. Genetics (2014). doi: 10.1093/hmg/ddu257.
HTML PDF
Bohic M, Marics I, De Pais Paiva Santos C, Ben-Arie N, Malapert P, Salio C, Reynders A, Le Feuvre Y, Saurin AJ, and Moqrich A.
Loss of bhlha9 impairs thermotaxis and formalin-evoked pain in a sexually dimorphic manner.
Cell Reports 30 (3): 602-610.e6 (2020).
HTML PDF
METHODS
Krizhanovsky V, Golenser E, and Ben-Arie N.
Genotype identification of Math1/LacZ knockout mice based on real-time PCR with SYBR Green I dye.
J Neurosci Methods 136: 187-192 (2004). http://www.ncbi.nlm.nih.gov/pubmed/15183270
Schatz O, Golenser E, and Ben-Arie N.
Clearing and photography of whole mount X-gal stained mouse embryos.
Biotechniques 39: 650-654 (2005). http://www.ncbi.nlm.nih.gov/pubmed/16312214
HTML PDF
Genotype identification of Math1/LacZ knockout mice based on real-time PCR with SYBR Green I dye.
J Neurosci Methods 136: 187-192 (2004). http://www.ncbi.nlm.nih.gov/pubmed/15183270
Schatz O, Golenser E, and Ben-Arie N.
Clearing and photography of whole mount X-gal stained mouse embryos.
Biotechniques 39: 650-654 (2005). http://www.ncbi.nlm.nih.gov/pubmed/16312214
HTML PDF
